Daisuke Wakabayashi and Cade Metz report that Google “has fired a researcher who questioned a paper it published on the abilities of a specialized type of artificial intelligence used in making computer chips”:
The researcher, Satrajit Chatterjee, led a team of scientists in challenging the celebrated research paper, which appeared last year in the scientific journal Nature and said computers were able to design certain parts of a computer chip faster and better than human beings.
Dr. Chatterjee, 43, was fired in March, shortly after Google told his team that it would not publish a paper that rebutted some of the claims made in Nature, said four people familiar with the situation who were not permitted to speak openly on the matter. . . . Google declined to elaborate about Dr. Chatterjee’s dismissal, but it offered a full-throated defense of the research he criticized and of its unwillingness to publish his assessment.
“We thoroughly vetted the original Nature paper and stand by the peer-reviewed results,” Zoubin Ghahramani, a vice president at Google Research, said in a written statement. “We also rigorously investigated the technical claims of a subsequent submission, and it did not meet our standards for publication.” . . .
The paper in Nature, published last June, promoted a technology called reinforcement learning, which the paper said could improve the design of computer chips. . . . Google had been working on applying the machine learning technique to chip design for years, and it published a similar paper a year earlier. Around that time, Google asked Dr. Chatterjee, who has a doctorate in computer science from the University of California, Berkeley, and had worked as a research scientist at Intel, to see if the approach could be sold or licensed to a chip design company . . .
But Dr. Chatterjee expressed reservations in an internal email about some of the paper’s claims and questioned whether the technology had been rigorously tested . . . While the debate about that research continued, Google pitched another paper to Nature. For the submission, Google made some adjustments to the earlier paper and removed the names of two authors, who had worked closely with Dr. Chatterjee and had also expressed concerns about the paper’s main claims . . .
Google allowed Dr. Chatterjee and a handful of internal and external researchers to work on a paper that challenged some of its claims.
The team submitted the rebuttal paper to a so-called resolution committee for publication approval. Months later, the paper was rejected.
There is another side to the story, though:
Ms. Goldie [one of the authors of the recently published article on chip design] said that Dr. Chatterjee had asked to manage their project in 2019 and that they had declined. When he later criticized it, she said, he could not substantiate his complaints and ignored the evidence they presented in response.
“Sat Chatterjee has waged a campaign of misinformation against me and [coauthor] Azalia for over two years now,” Ms. Goldie said in a written statement.
She said the work had been peer-reviewed by Nature, one of the most prestigious scientific publications. And she added that Google had used their methods to build new chips and that these chips were currently used in Google’s computer data centers.
And an outsider perspective:
After the rebuttal paper was shared with academics and other experts outside Google, the controversy spread throughout the global community of researchers who specialize in chip design.
The chip maker Nvidia says it has used methods for chip design that are similar to Google’s, but some experts are unsure what Google’s research means for the larger tech industry.
“If this is really working well, it would be a really great thing,” said Jens Lienig, a professor at the Dresden University of Technology in Germany, referring to the A.I. technology described in Google’s paper. “But it is not clear if it is working.”
The above-linked news article has links to the recent paper in Nature (“A graph placement methodology for fast chip design,” by Azalia Mirhoseini) and the earlier preprint (“Chip placement with deep reinforcement learning”), but I didn’t see any link to the Chatterjee et al. response. The news article says that “the rebuttal paper was shared with academics and other experts outside Google,” so it must be out there somewhere, but I couldn’t find it in a quick, ummmmm, Google search. The closest I came was this news article by Subham Mitra that reports:
The new episode emerged after the scientific journal Nature in June published “A graph placement methodology for fast chip design,” led by Google scientists Azalia Mirhoseini and Anna Goldie. They discovered that AI could complete a key step in the design process for chips, known as floorplanning, faster and better than an unspecified human expert, a subjective reference point.
But other Google colleagues in a paper that was anonymously posted online in March – “Stronger Baselines for Evaluating Deep Reinforcement Learning in Chip Placement” – found that two alternative approaches based on basic software outperform the AI. One beat it on a well-known test, and the other on a proprietary Google rubric.
Google declined to comment on the leaked draft, but two workers confirmed its authenticity.
I searched on “Stronger Baselines for Evaluating Deep Learning in Chip Placement” but couldn’t find anything. So no opportunity to read the two papers side by side.
Comparison to humans or comparison to default software?
I can’t judge the technical controversy given the available information. From the abstract to “A graph placement methodology for fast chip design”:
Despite five decades of research, chip floorplanning has defied automation, requiring months of intense effort by physical design engineers to produce manufacturable layouts. Here we present a deep reinforcement learning approach to chip floorplanning. In under six hours, our method automatically generates chip floorplans that are superior or comparable to those produced by humans in all key metrics, including power consumption, performance and chip area. . . . Our method was used to design the next generation of Google’s artificial intelligence (AI) accelerators . . .
This abstract is all about comparisons with humans, but it seems that this is not the key issue, if the Stronger Baselines article was claiming that “two alternative approaches based on basic software outperform the AI.” I did find this bit near the end of the “A graph placement methodology” article:
Comparing with baseline methods. In this section, we compare our method with the state-of-the-art RePlAce and with the production design of the previous generation of TPU, which was generated by a team of human physical designers. . . . To perform a fair comparison, we ensured that all methods had the same experimental setup, including the same inputs and the same EDA tool settings. . . . For our method, we use a policy pre-trained on the largest dataset (20 TPU blocks) and then fine-tune it on five target unseen blocks (denoted as blocks 1–5) for no more than 6 h. For confidentiality reasons, we cannot disclose the details of these blocks, but each contains up to a few hundred macros and millions of standard cells. . . . As shown in Table 1, our method outperforms RePlAce in generating placements that meet design criteria. . . .
There’s a potential hole here in “For confidentiality reasons, we cannot disclose the details of these blocks,” but I don’t really know. The article is making some specific claims so I’d like to see the specifics in the rebuttal.
It doesn’t sound like there’s much dispute about the claim that automated methods can outperform human design. That is not a huge surprise, given that this is a well-defined optimization problem. Indeed, I’d like to see some discussion of what aspects of the problem make it so difficult that it wasn’t already machine-optimized. From the abstract: “Despite five decades of research, chip floorplanning has defied automation, requiring months of intense effort by physical design engineers to produce manufacturable layouts,” but the article also refers to “the state-of-the-art RePlAce,” so does that mean that RePLAce is only partly automatic?
The whole thing is a bit mysterious to me. I’m not saying the authors of this paper did anything wrong; I just don’t quite understand what’s being claimed here: in one place the big deal seems to be that this procedure is being automated; elsewhere the dispute seems to be a comparison to basic software.
Google’s problems with reproducibility
Google produces some great software. They also seem to follow the tech industry strategy of promoting vaporware, or, as we’d say in science, non-reproducible research.
We’ve seen two recent examples:
1. The LaMDA chatbot, which was extravagantly promoted by Google engineer Blaise Agüera y Arcas but with a bunch of non-reproducible examples. I posted on this multiple times and also contacted people within Google, but neither Agüera y Arcas nor anyone else has come forth with any evidence that the impressive conversational behavior claimed from LaMDA is reproducible. It might have happened or it might all be a product of careful editing, selection, and initialization—I have no idea!
2. University of California professor and Google employee Matthew Walker, who misrepresents data and promotes junk science regarding sleep.
That doesn’t mean that Chatterjee is correct in the above dispute. I’m just saying it’s a complicated world out there, and you can’t necessarily believe a scientific or engineering claim coming out of Google (or anywhere else).
P.S. From comments, it seems that Google no longer employs Matthew Walker. That makes sense. It always seemed to a misfit for a data-focused company to be working with someone who’s famous for misrepresenting data.
P.P.S. An anonymous tipster sent me the mysterious Stronger Baselines paper. Here it is, and here’s the abstract:
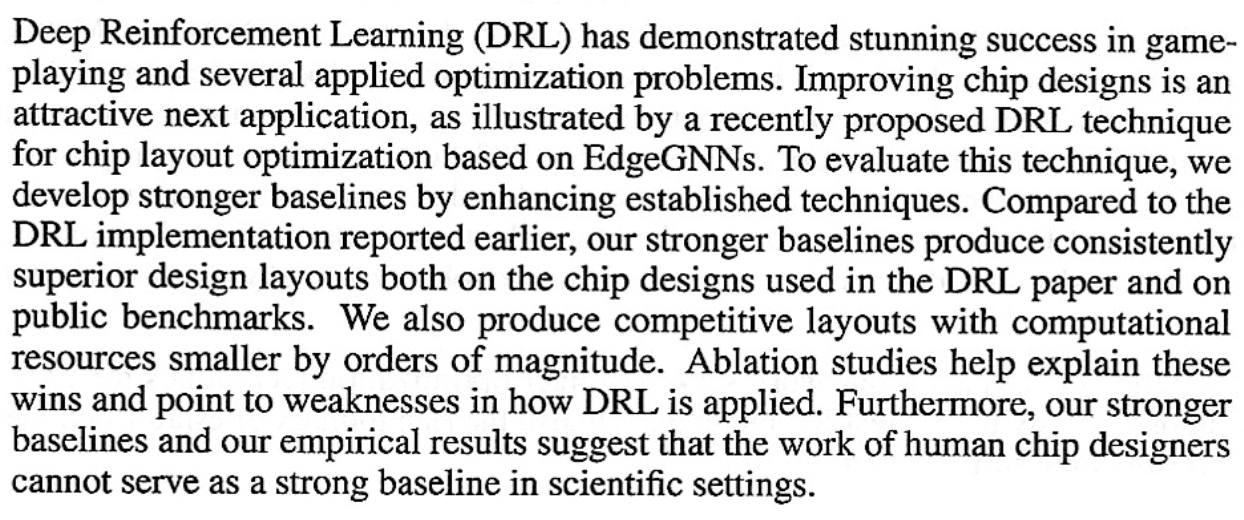
This all leaves me even more confused. If Chatterjee et al. can “produce competitive layouts with computational resources smaller by orders of magnitude,” they could show that, no? Or, I guess not, because it’s all trade secrets.
This represents a real challenge for scholarly journals. On one hand, you don’t want them publishing research that can’t be reproduced; on the other hand, if the best work is being done in proprietary settings, what can you do? I don’t know the answer here. This is all setting aside personnel disputes at Google.
See also: https://www.sciencedaily.com/releases/2020/10/201014114606.htm
I’ve seen it linked directly by somebody, but the pdf vanished when I checked it out again. Possibly it used some proprietary code or data without license, I can only imagine. There was no author in the pdf itself. But as you highlighted, the main point that gets obfuscated in the convoluted NYT article it’s whether the new method has consistent advantages over some off-the-shelf solutions. As an outsider to chip design with zero stakes at it, I can only think that this claim should be easy to check so to confirm/discard it – and if it’s not easy then how reliable is the tech after all?
Matthew Walker was at Google for a very short time, you might want to update on that.
Luiz:
Oh, I had no idea! Do you have a link for that?
His linkedin says he was a “Sleep Scientist advisor” from Oct 2019 to Feb 2020
https://www.linkedin.com/in/sleepdiplomat
Interesting story. Reproducibility problems with deep reinforcement learning have been talked about for a couple years. Joelle Pineau, for instance, who has organized some of the reproducibility challenges at ML conferences, noted a bunch of ways in which evaluations in deep RL are particularly prone to misleading results due to non-determinism in the mechanism (see eg https://www.youtube.com/watch?v=Vh4H0gOwdIg, https://ojs.aaai.org/index.php/AAAI/article/view/11694)
The more general problem with baselines not being appropriately defined or tuned has been coming up a lot lately across different areas of ML (we summarize some examples in our recent paper, which I will blog about one of these days: https://arxiv.org/pdf/2203.06498.pdf)
But yeah, too many holes in this story to say how much weight to put on the claims against the published paper.
Jessica:
Yeah, arguably the journal should just not have even considered publishing the paper, given this bit: “For confidentiality reasons, we cannot disclose the details of these blocks, but each contains up to a few hundred macros and millions of standard cells.”
There are lots of places where Google engineers can publicize their research. It’s not clear that scientific journals are the right place, if the work is not reproducible. It’s enough to say that the work is not reproducible, without taking any position on the criticism of that work.
No expert here. What follows is pure guesswork
1. (less important) layouts seem like a non-differentiable knapsack problem, so the straightforward optimization approach is probably np-hard and exponential in a massive number of node placements. From skimming the paper, it like they make it a differentiable optimization problem here by forming it into a sequence of individual placements with a reward function predicted by a neural network, then backpropagating the final chip reward through all the steps it took. That is, at each state, given a half-formed chip with node placements already made, the network places the next node to maximize expected cumulative reward. This was probably only a feasible approach relatively recently since the space of possible actions at every individual step is already massive, so we need big networks, many iterations, distributed training across many GPUs and low precision ASICs for fast evaluation, which are all relatively new.
2. (more important) I think the real killer here is “manufacturable”. While computationally evaluating the “quality” of a chip is straightforward, figuring out how expensive a given layout is to actually fabricate at scale seems pretty much undecidable. Fabrication technology is also changing every single year. In this paper, it looks like they only take into account hard constraints
But if I had to guess, soft constraints and scalable fabrication is the real killer here, and also what skeptics mean when they say “but it is not clear if it is working”. They do claim
but you know–only Google knows. There’s only a handful of institutions in the world that can conduct the full-stack of this kind of research, and they all have simultaneous incentives to keep the final product as a trade secret if it works and also to build hype anyways if it doesn’t.
After reading some more
1. It does seem like this is a non-differentiable NP-hard combinatorial optimization problem and can’t be attacked head-on, and that computational resources were and still are a problem even for heuristic methods like this one.
2. it doesn’t look like manufacturability is a real concern at this stage of design. Seems like any valid design can basically be printed.
The most interesting part of the problem here is–are these computer-optimized chips something we can actually make? The only way to figure that out is to make a serious effort at trying to make them, and the only people who can even attempt it are foundries (GlobalFoundries) and their customers (Google->Broadcom).
Not sure how long it will be there, but I found it here
http://47.190.89.225/pub/education/MLcontra.pdf
Some of the stuff in this paper makes no sense at all to me. For example, in section 3, Chatterjee claims that “the results in the Nature paper are noot well-defined” and “design quality attained by a human designer is a subjective baseline.” But, the Nature paper’s final results are a table of timing, congestion, and wirelength.
He also writes “the superiority of human designers over chip layout software has not been established in the Nature paper or prior publications. Moreover, by demonstrating software tools that outperform the novel RL technique, it would be possible to disprove this claim.” And in the abstract, he writes “our stronger baselines…suggest that the work of human chip designers cannot serve as a strong baseline in scientific settings.” But in the actual nature article, the “manual” layouts they’re comparing against are actually using a commercial EDA tool with a human in the loop.
so unless the expert chip designers are actually making things worse and also not checking for those regressions in between iterations, they’re at least as good as the software he claims is a strong baseline.
Also in the intro, he writes “Although there is a long history of applying AI and machine-learning techniques in EDA, only recently has revolutionary success–int he sense of an EDA system being ‘superhuman’ at solving a placement problem–been claimed in a scientific setting.” which seems to directly contradict his later assertion that “the superiority of human designers over chip layout software has not been established”.
Later, he criticizes the nature article for comparing and combining tools that optimize different objectives, since congestion is part of the reward function for the nature method but is not included in the existing tools for lack of convexity/differentiability. That doesn’t really make sense as a criticism to me (again, a complete layman) since the final comparisons are trying to get at performance. The ability to add a non-differentiable non-convex target to the reward function is pretty explicitly presented as a benefit in the original paper. The nature article also writes
Then in section 3.1 he claims “the objective for all the methods [which includes the nature method] is to minimize wirelength” because it’s “much faster to compute in an optimization loop” and “none of the tools explicitly optimize for congestion”. All that makes me think that he removed congestion from the reward mixture when running the reinforcement learning approach to try and make the comparison more fair and performant. Even in the original article, reinforcement learning underperformed the standard on wirelength. The only real empirical discrepancy in the results is that in the original article reinforcement learning improved overall compute performance and decreased congestion, while here it doesn’t.
Again, I’m a complete layman here, so it’s possible this is entirely misinterpretation on my part. But this rebuttal paper really doesn’t seem like it’s up to the quality of the original paper. It also seems like some of these claims can be reproduced or unambiguously resolved by a subject matter expert, since Chatterjee is at least right that his own experiments are more reproducible:
1. Does the google approach, performed as originally described, really underperform RePlAce on the IBM benchmarks?
2. Does a human in the optimization loop typically improve performance for commercial layout software or not?
Hi Andrew,
I think the NYT article is actually quite irresponsible in how it portrays the situation. My understanding is that Chatterjee was fired after a years-long campaign to undermine and harass the two lead authors of the paper. This isn’t a matter of shutting up criticism but rather one of workplace misconduct. (It’s also worth noting that Chatterjee was not an employee at Brain, so it seems rather unusual for him to ask to manage two Brain researchers.)
From having known Anna for a while, she’s one of the most impressive junior ML researchers I’m aware of, and always holds herself to a high standard of integrity. It’s pretty gross that the NYT decided to platform Chatterjee’s continued bullying of two junior researchers.
Best,
Jacob
Yikes. Good to know.
Jacob:
Thanks for sharing this information. I guess the story is difficult to handle because it has four aspects:
1. Scholarly journals publishing non-reproducible research.
2. Research criticism in a corporate environment.
3. High-tech developments in algorithms.
4. Workplace misconduct.
And the four parts of the story get tangled in the telling.
Thanks! I appreciate the response. What’s frustrating is that NYT went with the angle that would get the most clicks, without caring much about the consequences for those involved (many people will first hear about this paper through the NYT article). I’m glad that you are applying more nuance than the NYT did.
First time reading the NYT?
Agree; #2 seems most explanatory.
One additional comment: it looks like the code actually is open source: https://twitter.com/yablak/status/1521136538062524416. Someone else in that thread also claims that their team independently replicated the results (internally to Google).
Dear Jacob,
I am wondering how you could put all the pieces of this puzzle together this fast and jump to a conclusion (even before reading the rebuttal paper).
The open source code simply does nothing and its purely a marketing hoax, no release of the clustering algorithm, no release of SA, and only for one block (Ariane).
The open source claims that the hyper-parameters were “slightly” changed compared to the Nature paper:
“Some hyperparameters are changed from the paper to make the training more stable for the Ariane block.”
If you closely look, the changes in the hyper-parameters have nothing to do with the training stability and are purely related to the proxy cost function. Plus, a 10x increase in the proxy cost weights (density_weight from 0.1 to 1.0 and congestion_weight from 0.1 to 1.0) should not be considered a “slight” change.
While I understand that the release of the TPU blocks (Table 1 Nature paper) may not be possible due to the proprietary reasons, but I encourage the authors and the open source team to release the training curves (Tensorboards) for the TPU blocks in the paper, which I assume you would agree that is the bare minimum requirement for machine learning papers.
Chaterjee’s paper is convincing. It deserves publication if the original did. It seems he got on the wrong side of an organizational desire to oversell the org’s DL achievement.
If more scientists were willing to undertake such crusades, there would be more discord but also more accurate presentation of scientific knowledge + contributions to it.
We have no way of knowing (or reason to care) what level of personal conflict was involved to motivate him in this. Ideally the original paper would have included his considerations, but it seems from how this one went that Google publications are probably managed so as to maximize Google PR more than to help outside-Google researchers.
Thanks for this article, a platform for open discussion.
Jacob Steinhardt I have seen that you started a campaign in Twitter accusing/bombarding Sat based on heresy and attacking free journalism. I am curious to know whether you tried to read the rebuttal paper. If so, which parts of the paper doesn’t sound right to you?
Wouldn’t it be better to at least reach out to other party and hear their side? Interestingly, you answered at least one of the misunderstandings around this matter in one of your replies. How come someone from a different team can even request to mange people from a different team? This is an utterly false claim.
Wouldn’t it be better to attempt reproducing Nature results (which I doubt you can since everything is “proprietary”) instead of doing the easy thing and attacking others?
Aren’t you slightly curious that the “Electronic Design Automation (EDA)” community has raises multiple concerns around this work? See https://www.youtube.com/playlist?list=PL6-vor2YamEBF4cXQwfsgYJon4SPSMv0-
I give you one example of discrepancies in Nature results (there are more):
** February 20, 2020 **
TheNextPlatform Article: https://www.nextplatform.com/2020/02/20/google-teaches-ai-to-play-the-game-of-chip-design/
Results on a TPU Design Block
Time taken: 24 hours, Total wirelength: 55.42m
** November 03, 2020 **
Exact same results were presented in the Nature paper:
Extended Data Fig. 5 Visualization of a real TPU chip (same chip with an identical placement)
Time taken: 6 hours, Total wirelength: 55.42m
** November 17, 2021 **
Exact same results were presented in Chips Alliance: https://chipsalliance.org/blog/2021/11/17/how-google-is-applying-machine-learning-to-macro-placement/
Time taken: 24 hours, Total wirelength: 55.42m
I took some time to read the leaked Stronger Baselines paper and have a question: why does the rebuttal not mention the hyperparameters used in the RL experimental setup? RL requires a careful choice of hyperparameters to perform well, and if Chatterjee et al. are truly with good intent then they should have released their hyperparameters. Any ML conference would likely reject this paper due to this obvious omission, and given how obvious this is, one might suspect that this omission was deliberate.
That’s not a discrepancy. 24 hours is “from scratch”, 6 hours is just time spent “fine tuning”, meaning taking a pretrained model from a more general problem and training it some more on a specific problem.
I’m curious what other discrepancies there are and whether or not they are errors of reading comprehension.
I can’t find the community consternation in your YouTube link, it’s just a link the the whole symposium.
The NYT article was pretty much devoid of technical content, and focused on the workplace drama angle. My replies here and on twitter were addressing the article on that level. I fully support academic criticism in general, and of this paper in particular, but it’s a huge stretch to call the NYT article a piece of academic criticism. Moreover, the press isn’t the right venue for academic discussion–blogs such as Andrew’s are much better, and I haven’t objected to any of the discussion here.
I haven’t yet read the rebuttal, although I hope to soon. It seems unreasonable to insist that I digest a 12-page paper before contributing other true and relevant information about the situation. This story is now fast-moving (Chaterjee’s decision by going to the Times), and I wanted to correct the record quickly. I’m not sure what you mean by attacking free journalism–it’s my duty as a citizen to correct the press when they make mistakes.
Anyways, I look forward to a discussion on the academic merits of the paper. I’d personally be pretty surprised if their main claims didn’t hold up, given the results were independently reproduced by a second team at Google and are used in production for TPU design. But I’m always open to information and arguments.
(sorry, the above was meant to be a reply to the commenter “somebody”. not sure how to edit/delete comments)
Hey, I didn’t say that! That was “someday.”
I’m guessing it’s Chaterjee actually
It is not clear what you are trying to say, Jacob. The NYT article discusses how Google promotes its non-science in a fascist manner. Your position seems unreasonable. It makes me wonder whether you have ties to Farinaz Koushanfar or her gang member Tara Javidi. Your Twitter campaign is disgusting and utterly stupid. You should be ashamed of yourself. I think you are more of a 1%-er wannabe than NYT is the establishment. How can you be tenure-track at Berkeley? Oh, wait, just like your friends, through a mafia (a.k.a illegitimate academic network).
Cyrus:
Hey, let’s go easy on the personal attacks here.
I don’t know the details, but “undermine and harass” could cover a lot of different actions. Do you know what precisely misconduct Chatterjee was accused of doing? If it’s just, “he just criticizing our work, even after Google said his work doesn’t meet its standards for publication,” that might be getting into “methodological terrorist” terrritory, depending upon precisely what he did. I could definitely see how persistent criticism could cross a line into harassment. Still,
I guess the wider issue that’s followed in the wake of the replication crisis is how we distinguish critique that persists against establishment attempts to keep criticism in-house (either within Google, PNAS, etc.) and behaviour that constitutes actual harassment. “Bropenscience” is obviously something we should all be concerned about, but it is also a charge that can be conveniently thrown around to dismiss legitimate critique, along with “methodological terrorists” and “online vigilantes”.
(I suspect that most researchers would be able to find someone who would say about them, “I know X personally and they have a high standard of intellectual integrity so these repeated critiques of their work is just bullying”).
Thanks Jacob for the reply. I don’t want to dive into the details of your tweet, but you started by calling Sat a bully and using such terms as “in reality…”– I am not sure how you came to such a strong conclusion.
I had respect for Timnit, but after observing her immediately making this a gender matter without even a shred of evidence, she lost me right there. I agree with your statement about Google having a problem with Sat; that problem is him speaking up to set the science straight. If it turns out that he did something remotely inappropriate, I will be the loudest voice in the community.
Unfortunately, the environment is so heated that it is hard to have an open discussion.
Let’s continue the technical discussion about the paper and the reproducibility of the results.
The github does not reproduce the Nature paper. It only provides the code for one public block, while not releasing the code for “Simulated Annealing” (also closely observe the open issues on that github). I am curious to know whether the authors were aware of the new SA moves presented in the Stronger Baselines paper (Table 2) —which provide significantly better results compared to what was presented in the Nature paper— before their publication and if so, why did they decide to not include those SA moves (favoring RL)?
Please consider that there is at least a small chance of scientific misconduct.
“The methodology from the Nature paper” that means the exact same parameter from Nature paper. Are you saying that for each design the Nature paper should tune the hyper-parameters? So it is not 6 hours? Interesting!
No. Not hyperparameter tuning. Training. This is a very common workflow in deep learning, it’s called transfer learning and it’s written about clearly in the paper. The time spent on the problem is indeed 6 hours. However, they’re starting from a model that has already been trained on more general problems for much longer. If you include both training times, then indeed it’d be much longer than 6 hours. However, the idea is that you only have to train the general model once. It’s the same exact idea as GPT-3. Train a big general model slowly once, then you can attack many problems with less training.
But the point is that your “discrepancy” of 6 hours vs 24 hours is just an error in your own reading comprehension. They state clearly in the paper that 24 hours is for a model trained “from scratch”, and that the from scratch approach is not the one they went with for their final results.
To be clear, I’m generally somewhat of an AI and deep learning skeptic. I also agree that the nature paper is not reproducible, and its publication in a journal is of questionable value. But it really does feel like your “discrepancies” are the result of not actually reading the paper.
@someday: This is not an actual quote from the paper, I looked through the Stronger Baselines paper just now and couldn’t find it anywhere. Instead, it states in Section 3.1 Experimental Setup: “For the two-step methodology from Nature, we use the (code of the) RL implementation described in the Nature paper to place macros”.
Using the code of the RL implementation does not include the hyperparameters, since hyperparameters are obviously not code. How can we trust you when you’re making up quotes from thin air? Interesting indeed!
Please read the paper instead of searching for words. The quote is from page 4, Sec 3.1. Also, read experimental setup.
I’ve already quoted to you what it says in Sec 3.1 Experimental Setup. It states that you used only the code of the RL implementation, not necessarily the same configuration including hyperparameters.
I’ve re-read the entirety of Section 3.1. Here’s the full context: “Two step: The methodology from the Nature paper that first clusters the circuit, then places macros, then places smaller circuit components (standard cells)”. This doesn’t mean that the same hyperparameters were used, just that the [clustering -> macro placement -> standard cell placement] approach was used.
It is standard practice to provide hyperparameters due to their importance towards ML training convergence. Please provide the exact hyperparameters used in your experiments.
Well first you said you can not find the quote and now something else.
To my understanding, “using the same codebase” and “The methodology from the Nature paper” are self-explanatory, but if you are this deeply caught up in this, you can reach out to authors or your friends at Google to check out the code for you.
Also, ask your friends about “Experiments for Nature rebuttal” document, that would help you to discover the mysteries.
I have consistently said that you (I assume you’re Satrajit Chatterjee based on the way you’re acting) didn’t provide the hyperparameters in the paper. My stance has not changed.
Neither “methodology” nor “(code of the RL) implementation” includes hyperparameters. Your story would be more credible if you provided hyperparameters, which is considered standard practice for any ML or RL paper. The fact that this is missing reflects poorly on you – especially since you supposedly have an ML background. Adding hyperparameters not only helps with replicating your results, but also can help demonstrate that you’re not intentionally handicapping RL algorithm performance by selecting bad hyperparameters.
You’ve mentioned “Experiments for Nature rebuttal” several times now, which has piqued my curiosity. Can you provide some context? What is this document and where is it located? Which part of the document would be interesting? What exactly would someone be looking for within the document?
I am not the author of any of the papers, so you can make your assumption in any way you want and I am not sure why you talk this aggressive. I answered to your question out of respect for the scientific community and this is the last reply.
My stance is also the same, based on what I read from the paper, I came to this conclusion that they used the exact same hyperparameters. I consistently requested you to email one of the authors for clarification and yet you ask here about hyperparameters.
For the other document, I also suggest you to contact one of the Nature’s authors. They can help you.
It’s highly unlikely that you’re not Satrajit Chatterjee or one of the other authors on his paper. Multiple people in this blog post (besides me) have already commented about your bizarre nitpicking behavior – and at least one other person has also identified you as Chatterjee.
You specifically asked above “which parts of the paper doesn’t sound right to you?” I am answering your question.
You have thanked this blog for providing “a platform for open discussion.” I’m providing the open discussion that you desired, yet you cannot even handle such a basic question about hyperparameters. I have more follow-up technical questions, but we can’t even get past the first, most basic question. Not a good sign.
Now, you’re asking that I email one of the authors. The authors of the Stronger Baselines rebuttal are redacted. If the authors truly stand by the contents of the paper, then why did they hide themselves from the paper? How can I contact the authors when they did not provide their contact information? Who are the authors?
Similarly, you ask that I find a strange “Experiments for Nature rebuttal” document. You implied that this is a Google-internal document. If you are not Chatterjee or one of the co-authors, why do you know about the existence and contents of a Google-internal document? What information do you think it contains? Your behavior is very strange indeed – why not just say what you actually mean instead of forcing us to guess?
I am glad that the authors are here. And yet reaching to exact same solution? Not only that, reaching to exact same placement?!!
Watch 4 3 @4:30
Watch 4 1 @7:30
Sorry it seems the replies are messed up.
My response was for this comment “RL requires a careful choice of hyperparameters to perform well”
> Are you saying that for each design the Nature paper should tune the hyper-parameters? So it is not 6 hours? Interesting!
Thanks for your comments and clarifications. I am familiar with the paper and read both Nature and stronger baseline. However, I think you are missing the point here. It is hardly unlikely to reach to the exact same outcome with transfer learning (not to mention even exact same placements — please look at the blurry placement figures and wirelength value). This is called random initialization in deep learning workflow.
Wait, so you’re claiming that the placements being the same across different press releases is evidence of misconduct?
This suggests an interesting issue related to publishing non-reproducible research. Here, the work is not reproducible for at least two reasons: 1) trade secrets; and 2) only certain people would have the resources to reproduce even if things were not secret.
Although #1 seems rather specific to this kind of research, it is a reasonable concern in most clinical work or work with human subjects. We intentionally obscure some information that might actually be necessary for replication (e.g., identifying combinations of demographic info) in the interests of privacy of our participants. This seems reasonable to me, and also if a result can only be reproduced in a population with very narrowly defined characteristics (e.g., only Cornell undergrads in 2010 have ESP, the rest of us are out of luck) then maybe it is not such a useful result after all.
#2 seems to be a pretty general concern. Only people with access to materials, equipment, computing resources, participants, etc., will be able to do research in the first place. And some research is necessarily more resource-intensive than others.
Finally, it strikes me that lots of fields by necessity do research that is not replicable in some sense. A lot of work in astronomy, paleontology, archeology, sociology, cosmology, anthropology, geology, etc., cannot be “reproduced” because it focuses on specific events, historical periods, societies, etc. that will never recur in the same form. That said, one could think of these issues as “in-principle replicable” in the sense that someone else with the same tools would have observed the same things and come to similar conclusions, it’s just that only certain people actually did so. And even if the specific circumstances never exactly recur, maybe we can replicate the features that we consider important, or use models.
Anyway, I guess this is all to say that it seems a lot of valuable research may be “unreplicable”, but there might be better or worse reasons.
A lot of criticism is directed to Matthew Walker at UC Berkeley. I think he’s acknowledged some of the problems in his book: https://sleepdiplomat.wordpress.com/2019/12/19/why-we-sleep-responses-to-questions-from-readers/.
Isn’t the point that everyone makes mistakes, but the important thing is to acknowledge them? If so, I’m not sure what else is expected.
Anon:
But Walker did not acknowledge the criticisms in any serious way. Indeed, in that post he neither mentions Guzey nor responds to Guzey’s specific points. Nor did Walker address his misrepresentation of the scientific literature.
Guzey discusses problems with Walker’s post here.
Who said misconduct? It is a discrepancy. And yes if you claim that you are generating the exact same placement with and without transfer learning, it needs explanation. For additional discrepancies, I would suggest you to read a document called “Experiments for Nature rebuttal”… I am positive you can find it. It was a great discussion. Good luck!
Dude, those are just different press releases of the same experiment. Why would they be different?
Ah, now I see what you’re talking about. The image in the early press release implies that it was the non-transfer learning run, but the same image in the paper implies that it was the transfer learning run. Was confused since that image isn’t in the arxiv version of the paper, I had to pirate it from sci-hub to see that. That is indeed a discrepancy, but frankly I don’t find it that suggestive. A search for this
“Experiments for Nature rebuttal”
comes up pretty empty for me though.
The Nature paper has been suspicious to me since the beginning. I am reasonably confident in my understanding of this topic to comprehend the two scientific papers and make my own judgement.
I agree with the point that scientific merits should be neither established nor defined in social media. To be honest, neither the NYT article nor the accusation campaign against one person mean much. If there is clear evidence of harassment, it should be public and potentially pursued via legal channels. If I were the authors, I would have kept the discussion technical and pursued the harassment allegations in court. I would personally support them during the legal process, because I believe EVERYONE should be heard and the US legal system is no cakewalk.
Let me get a little into the technical part of the work (I hope someone could share this broadly, believe me, not everyone has bad intentions). Just some requests for the authors.
Social media went crazy about the open source. This open source (at least this version) does not address the questions surrounding the reproducibility of the Nature paper. With all due respect to the ML community, EDA has been around for multiple decades, and the chip design flow consists of multiple processes (with their own noises) and different optimization goals and many nuances. For example, the last step in `Extended Data Fig. 1` in the Nature paper by itself is a non-deterministic process. That means, running PlaceOpt on two different designs could actually generate chips with similar quality (quality is also very subjective here — a design with slightly worse QoR could be easily favored over another one). Happy to elaborate more if needed. The point here is that proxy cost does **not** necessarily correlate to final QoR.
A major point that is missing in all these discussions is the claim about the superiority of machine learning/reinforcement learning over conventional methods (e.g. simulated annealing). “Extraordinary claims require extraordinary evidence.”
I am not going to question the results for now, but I believe the line between scientific breakthrough and marketing strategies is blurry. I could buy that a hand-tuned and significantly engineered machine learning method could perform optimization and finally turn out into a TPU chip, but I hardly believe that machine learning is at a stage to outperform traditional methods in EDA. I can be proven wrong with evidence and rigorous benchmarking and evaluations.
I sympathize with the authors as well. This is a hard time for them. I hope with more transparency, openness, and patience, we all can help move the science forward in a way that benefits everyone.
**Requests for Authors**
(1) Extended Data Table 4 presents some important results about the correlation between congestion weight and post-PlaceOpt performance. I am very surprised by these results (back to my discussion about noise in the process). Can you please extend this table with some other congestion weights (0.02, 0.05, 0.2, 0.5, 0.8, …)? Intuitively, there should be a turning point, post-PlaceOpt congestion can not decrease this nicely with congestion weight.
(2) Extended Data Table 6: it would be good to populate Table 1 with post-PlaceOpt results of simulated annealing. I assume in both SA and RL, you will use the exact same optimization objective with same weights and apply post-processing optimization to both cases, if any. Again refer to my point about noise. The intermediate proxy cost can not be the sole indicator of the final post-PlaceOpt performance.
(3) While I understand that releasing TPU blocks may not be possible (to be honest, I think they can anonymize the blocks and just release them, but it is ok, they are Google and I guess they can make these extraordinary claims), can you release the Tensorboards or training curves for these experiments? Especially, for RL results in Table 1 and SA results in Extended Data Table 6.
(4) Extended Data Table 5: you showed that there is limited sensitivity to the random seed choice. This is frankly shocking. Reviewing the large body of RL work, RL is inherently noisy and high variance (See PPO paper Figure 3, https://arxiv.org/pdf/1707.06347.pdf). Are there any algorithmic contributions here to prevent this inherent noise? The other possible explanation for these results is that they are obtained at different epochs/training time. Can you release the associated training time for each row?
(5) Somewhere in the paper the authors mentioned 20-30 TPU blocks. I am curious to see the performance of RL on all the blocks. Are there any limitations to the Nature method? I can see that in the review file, the authors mentioned “our method has produced optimized placements on every chip that we’ve tried it on so far”, this is one of those extraordinary claims in my opinion. Does it mean that for all the 20-30 TPU blocks, the Nature method was used for the final chip?
I would be happy to discuss these points further.
I am in the same camp. I am reasonably well versed in this field for both EDA and ML too.
The nature paper is an extraordinary claim and a little bit overstated (to mix a floorplan or more specifically macro placement problem as chip design).
Chip design is much longer process from architecture, RTL to implementation. Macro placement is just one step of many steps of chip design implementation. But all said, it is a decent work.
Look at the rebuttal work, if the result is reproduceable, it should at least allow to be published. it is known the ML vs human baseline maybe a moving line. Especially here is RL vs Human using EDA tools. The EDA tool itself have tons of knobs to tune, this is why we have to develop a AI tools to control the EDA tool parameters (like SNPS DSO.ai) and EDA tool is not static too. There is huge noise range from the human baseline part. How hard you have tried to use the EDA tool, how good the engineers on using the EDA tool matters too.
To me, the rebuttal will not reduce the value of the Nature paper but disallowing to publish do.
Just my 2 cents
Except that RL not only uses the PlaceOpt feedback for multiple months but also uses the human feedback! The Nature paper should just transparently explain the process, engineering effort, tuning, and the feedback they got from human and EDA, etc. Someone without EDA knowledge may think RL magically finds the placement under six hours as a push-button solution.
It is time AI conferences started introducing artifact badges. Many AI papers show improvement over SOTA in the first or second decimal place. It is hard to discern what is real and what is just an artifact of the randomness in the experimental evaluation.
+100
Has anyone noticed that the CPU usage for SA is different between the Nature (2 cpu) and the rebuttal (1 cpu)? Which one is right?
I read the paper ‘Stronger Baselines for Evaluation Deep RL in Chip Design’ and I have to say that the current state of the art for chip placement makes a complete mockery of the RL method. Operations Research for the win yet again.
Seriously though, why was that nature paper even celebrated?. I guess it was the title that made people lose their minds.
OTHO, Guzey continues to not respond to a number of criticisms of his own little grade school experiments and wild claims based on them. I feel kind of embarrassed for you whenever you point to his “work”.
Anon:
1. As a former grade schooler myself, I don’t like the use of “grade school” as an insult. Guzey is a guy in his 20s without funding to do sleep research. He’s transparently reporting what he’s done, and you and others can feel free to express your skepticism about it. Indeed, I myself expressed three points of skepticism in my post about his claims.
2. To continue on this point, it’s possible to express both open-mindedness and skepticism at the same idea. My friend Seth Roberts did lots of iffy experiments, and reported on them while expressing skepticism.
That said, I agree with you that Guzey (and, before him, Roberts) could do better at considering arguments that are critical of their theories. Speaking in general terms, it’s often difficult for people to criticize their own work or to respond well to outside criticism. That’s one good reason for the process of scientific discovery to be divided among multiple people. If Guzey publishes his work in a journal, then the journal should be open to publishing criticism of that work, or else another journal should publish relevant criticism. I posted on Guzey’s ideas on our blog, and I and others expressed skepticism and offered critical thoughts in the comments section. That’s how it goes. If Guzey could join in the discussion, that would be even better, but the participation of the original researcher is not required to have a useful exchange of ideas.
3. If you feel kind of embarrassed for someone else, that’s your problem, not theirs.
In this entire thread, I see how ignorant the ML/AI community is about chip design, and they just run in circles with their ignorance. Stop using the term chip design, and use macro placement instead.
I would like to make a general comment here. Jeff Dean should be questioned here, not a PhD student (Anna Goldie) and a junior Googler (Azalia Mirhoseini). They were persuaded to use fancy terms and sell a decade-old macro placement problem as a breakthrough in chip design by Jeff Dean! Jeff Dean’s multimillion-dollar compensation package is at stake here, which is why Google fired Sat. These kids are just tools and wannabe 1%-ers.
A superhuman macro placer, where you can just take the results as-is without mucking with them, and put them into a real chip, is definitely not a decade-old technology. Macro placement is a 60-year-old problem, but this is the first automated solution that outperforms and obviates the need for human experts.
This is absolutely a breakthrough, and I think represents the first time that RL has been used to solve a real-world engineering problem with actual economic value.
Plus, automating this step of the chip design process lets you automate upstream tasks.
Anna and Azalia are both Staff Research Scientists (one level above Senior Research Scientist). They are only junior researchers in the sense that they are not yet faculty, and so spurious rumors can have outsized impacts on their academic careers.
Non sense argument mostly. Macro placement is seldom done by humans in most design houses these days. Only incremental changes involve human expertise nowadays.
Dear Jacob. This is about science and not about people. Science is not rumor. Closing your eyes to facts does not solve anything. After one year Google still has not provided a robust platform to reproduce the results, to an extent that Andrew Kahng wrote a document and started to implement the code with his team.
https://docs.google.com/document/d/1vkPRgJEiLIyT22AkQNAxO8JtIKiL95diVdJ_O4AFtJ8/edit
Look there is no ending to this story unless Google supports its claims with transparency and true reproducibility. I don’t think anyone’s reputations are going to be ruined if all the supporting evidence and results are released. I would suggest you not to close your eyes to facts. I understand that they are your friends and you believe that you are supporting them, but the best help would be to advise them for more transparency.
I think the most correct statement in your comment is regarding the 60 year old history of macro placement.
If the Nature paper is ‘absolutely a breakthrough,’ then the authors of “Optimization by Simulated Annealing” should win the Turing Award. Don’t mistake Google-generated hype for a scientific breakthrough.
As for the authors’ career stage, what does this have to do with the validity of their methods? Mistakes can be committed by people at any level, and should be rectified. Whether one is a Staff Research Scientist or a postdoc or the God of RL, they should be held to the same standards.
Is something this application focused really a good fit for peer reviewed publications? Ultimately there’s a fairly limited set of companies that do chip design. The relevant question here is not whether Google’s method is better than google’s implementation of some existing method, but whether the new method beats or can beat those companies’ existing methodologies and thus is commercially viable or not.
https://medium.com/@mlgit_70611/bursting-the-bubble-a-case-against-reinforcement-learning-for-macro-placement-1c0d014c4143
Update: The researchers’ code was open-sourced: https://github.com/google-research/circuit_training
This repository has been up for over a year, right? Have things changed since this statement from Andrew Kahng? https://docs.google.com/document/d/1vkPRgJEiLIyT22AkQNAxO8JtIKiL95diVdJ_O4AFtJ8/edit
Relevant quote:
“Further, I believe it is well-understood that the reported methods are not fully implementable based on what is provided in the Nature paper and the Circuit Training repository. While the RL method is open-source, key preprocessing steps and interfaces are not yet available. I have been informed that the Google team is actively working to address this. Remedying this gap is necessary to achieve scientific clarity and a foundation upon which the field can move forward.”
Update: Prof. Kahng’s group came out with a paper in which, (if my reading of it is right) he says in very uncertain terms that the claims made my google in their nature paper was misleading.
here’s the paper: https://vlsicad.ucsd.edu/Publications/Conferences/396/c396.pdf
here’s his talk on it: https://vlsicad.ucsd.edu/Publications/Conferences/396/396.mp4
A rebuttal from the Google authors:
https://regmedia.co.uk/2023/03/27/ag_az_statement.pdf
Which the MacroPlacement repo seems to address on a point-by-point basis under “New FAQs” in their README:
https://github.com/TILOS-AI-Institute/MacroPlacement
This whole thing is… exhausting
Lawsuit from Chatterjee with 40 pages on one side of the story: https://regmedia.co.uk/2023/03/26/satrajit_vs_google.pdf