Andrew and I both received a note like this from NVIDIA:
We have reviewed your NVIDIA GPU Grant Request and are happy support your work with the donation of (1) Titan Xp to support your research.
Thanks!
In case other people are interested, NVIDA’s GPU grant program provides ways for faculty or research scientists to request GPUs; they also have graduate fellowships and larger programs.
Stan on the GPU
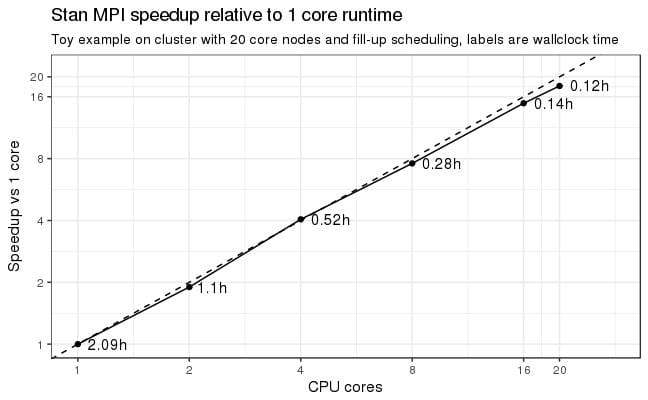
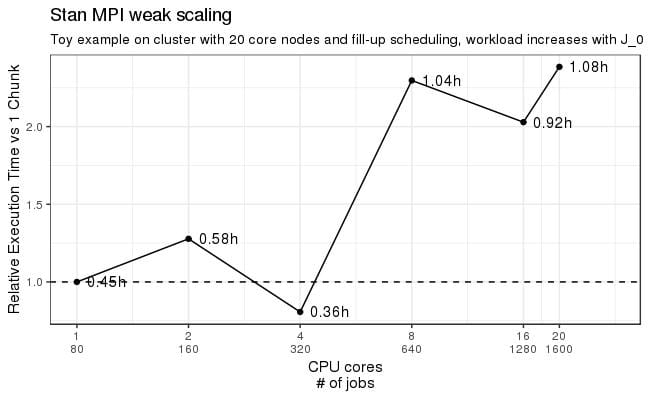
The pull requests are stacked up and being reviewed and integrated into the testing framework as I write this. Stan 2.19 (or 2.20 if we get out a quick 2.19 in the next month) will have OpenCL-based GPU support for double-precision matrix operations like multiplication and Cholesky decomposition. And the GPU speedups are stackable with the multi-core MPI speedups that just came out in CmdStan 2.18 (RStan and PyStan 2.18 are in process and will be out soon).
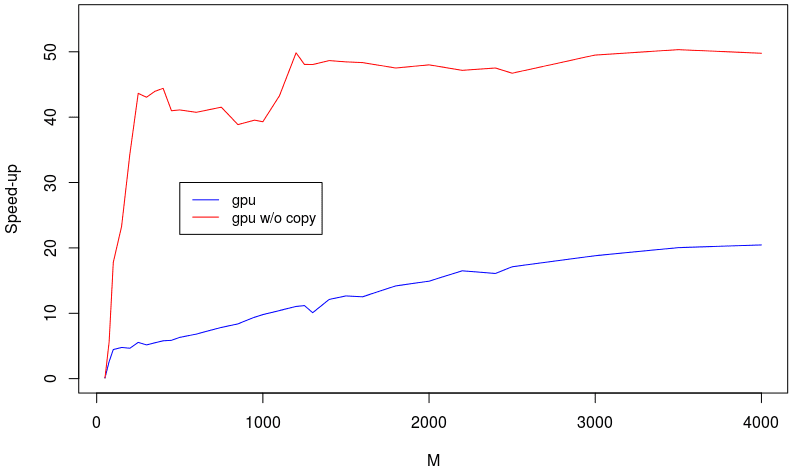
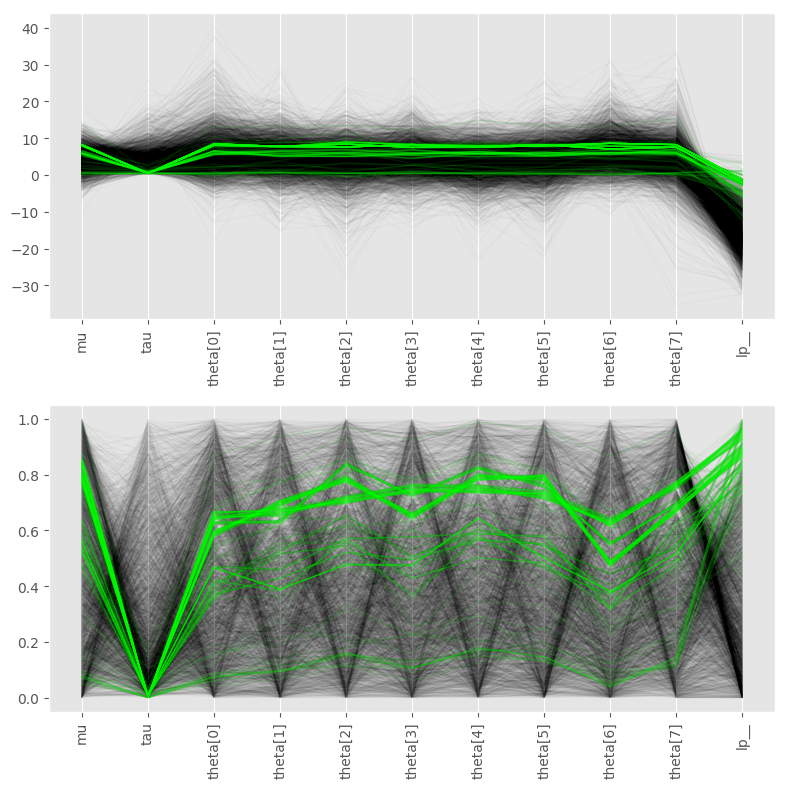
Figure 1. The plot shows the latest performance figures for Cholesky factorization; the X-axis is the matrix dimensionality and the Y-axis the speedup vs. the regular Cholesky factorization. I’m afraid I don’t know which CPU/GPU combo this was tested on.
Academic hardware grants
I’ve spent my academic career coasting on donated hardware back when hardware was a bigger deal. It started at Edinburgh in the mid-80s with a Sun Workstation I donated to our department. LaTeX on the big screen was just game changing over working on a terminal then printing the postscript. Then we got Dandelions from Xerox (crazy Lisp machines with a do-what-I-mean command line), continued with really great HP Unix workstations at Carnegie Mellon that had crazy high-res CRT monitors for the late ’80s. Then I went into industry, where we had to pay for hardware. Now that I’m back in academia, I’m glad to see there are still hardware grant programs.
Stan development is global
We’re also psyched that so much core Stan development is coming from outside of Columbia. For the core GPU developers, Steve Bronder is at Capital One and Erik Štrumbelj and Rok Češnovar are at the University of Ljubljana. Erik’s the one who tipped me off about the NVIDIA GPU Grant program.
Daniel Lee is also helping out with the builds and testing and coding standards, and he’s at Generable. Sean Talts is also working on the integration here at Columbia; he played a key design role in the recent MPI launch, which was largely coded by Sebastian Weber at Novartis in Basel.