In Bayesian statistics one often has to justify the prior used. While picking the prior may be seen as a subjective preference, if a different person disagrees with the prior, the conclusions would not be acceptable. For the results of the analysis to be acceptable, a large number of people would need to agree with the prior. Priors are not particularly easy to understand and evaluate, especially when the models become complex.
On the other hand, the fundamental idea of data splitting is very intuitive: we hide one part of the data, learn on the rest, and then check our “knowledge” on what was hidden. There is a hidden parameter here: how much we hide and how much we show. Traditionally, “cross-validation” meant the same as is currently meant by “leave-one-out”: we hide a single case, model from all the others, and then compute the error of the model on that hidden case.
As to remove the influence of a particular choice of what to show and what to hide, we ideally average over all show/hide partitions of the data. The contemporary understanding of “k-fold cross-validation” is different from the traditional one. We split the cases at random into k groups, so that each group has approximately equal size. We then build k models, each time omitting one of the groups. We evaluate each model on the group that was omitted. For n cases, n-fold cross-validation would correspond to leave-one-out. But with a smaller k, we can perform the evaluation much more efficiently.
When doing cross-validation, there is the danger of affecting the error estimates with an arbitrary assignment to groups. In fact, one would wonder how does k-fold cross-validation compare to repeatedly splitting 1/k of the data into the hidden set and (k-1)/k of the data into the shown set.
As to compare cross-validation with random splitting, we did a small experiment, on a medical dataset with 286 cases. We built a logistic regression on the shown data and then computed the Brier score of a logistic regression model on the hidden data. The partitions were generated in two ways, using data splitting and using cross-validation. The image below shows that 10-fold cross-validation converges quite a bit faster to the same value as does repeated data splitting. Still, more than 20 replications of 10-fold cross-validation are needed for the Brier score estimate to become properly stabilized. Although it is hard to see, the resolution of the red line is 10 times lower than that of the green one, as cross-validation provides an assessment of the score after completing the 10 folds.
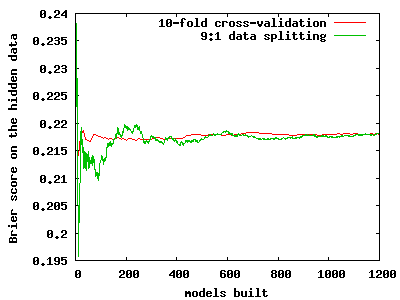
The implication is that the dependence of folds within a cross-validation experiment is a good thing if one tries to quickly assess the score. This dependence was seen as a problem before.
[Changes: the new picture does not show intermediate scores within each cross-validation, only the average score across all the cross-validation experiments.]
Cross-validation has been discussed earlier on this blog.